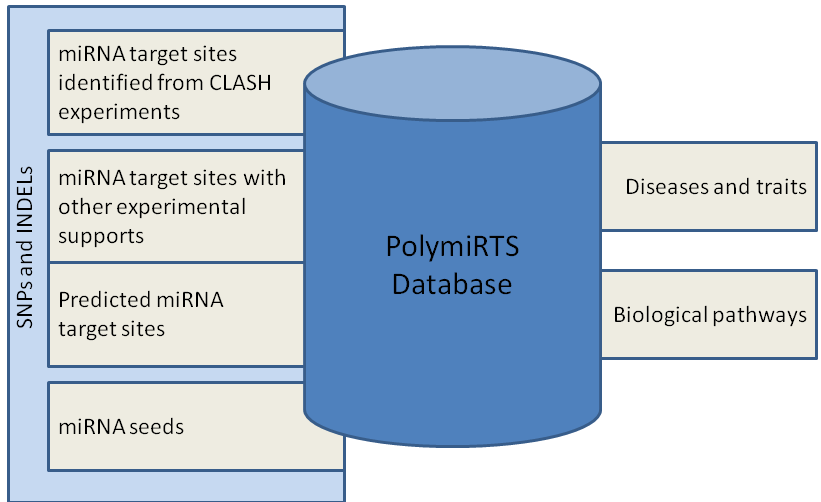
| PolymiRTS (Polymorphism in microRNAs and their TargetSites) is a database of naturally occurring DNA variations in microRNA (miRNA) seed regions and miRNA target sites. MicroRNAs pair to the transcripts of protein-coding genes and cause translational repression or mRNA destabilization. SNPs and INDELs in miRNAs and their target sites may affect miRNA-mRNA interaction, and hence affect miRNA-mediated gene repression.
PolymiRTS as genetic variants underlying variation in gene expression and disease phenotypes. Linkage analysis and association studies have long been used to discover chromosomal intervals harboring sequence variants that cause individual differences in biological traits. Polymorphisms in miRNA target sites are one type of sequence variant that cause these effects. The PolymiRTS database was created by scanning 3'UTRs of mRNAs in human and mouse for SNPs and INDELs in miRNA target sites. Then, the potential downstream effects of these polymorphisms on gene expression and higher-order phenotypes are identified. Specifically, genes containing PolymiRTSs, cis-acting expression QTLs, and physiological QTLs in mouse and the results of genome-wide association studies (GWAS) of human traits and diseases are linked in the database. The PolymiRTS database also includes polymorphisms in target sites that have been supported by a variety of experimental methods and polymorphisms in miRNA seed regions. |