Overview
SomamiR DB contains the somatic mutations that altering miRNA sequences and miRNA target sites in mRNA, lncRNAs, and circRNAs. We annotate these somatic mutations by linking them to a large collection of GWAS, CGAS, KEGG pathways. A newly developed web server miR2GO is integrated for the functional analysis of somatic mutations in miRNA seeds.
Somatic mutations that alter miRNA target sites
Clicking on the transcript ID from browse and search page will display all the somatic mutations in the target sites of miRNAs along with the most up-to-date collection of genetics and genomics data describing the functional impacts. Specifically, each transcript page contains the following information, if available:
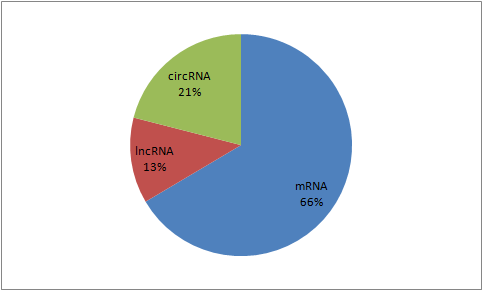
Somatic mutations on experimentally identified miRNA targets.
A. Experimentally identified miRNA target sites: CLASH
Cross linking, ligation, and sequencing of hybrids (CLASH) is a newly developed technique for high throughput mapping of RNA-RNA interactions that has recently been used for direct observation of miRNA-RNA target pairs associated with human AGO1. CLASH provides chimeric reads of miRNA and target site sequences and, therefore, directly identifies the targeting miRNA and allows for improved determination of non-canonical targeting. The CLASH dataset contains high-confidence canonical and non-canonical target sites of 399 different miRNAs.
B. Experimentally identified miRNA target sites: PAR-CLIP and HITS-CLIP
In addition to CLASH, several other CLIP-seq experimental techniques, such as the HITS-CLIP and PAR-CLIP have recently been developed and used to identify the specific mRNA sequences that bind with miRNAs in the RNA-inducing silencing complex (RISC). Unlike CLASH, these CLIP-seq data only gives the miRNA target sites. We identified all somatic mutations in the target sites that create, disrupt, or modify a 6mer or longer sequence complementary to a miRNA seed from two miRNA target predictions. First, we applied the 6 classes of miRNA seeds complementarity search as described by Ellwanger et al (PMID: 2144577). Second, we performed TargetScan prediction. By default, only the impacts of somatic mutations on target sites that are identified by TargetScan are shown. The impact of somatic mutations on all 6mer or longer seed matches can be displayed by clicking the "All 6mer or longer target sites" button on the top of the page.
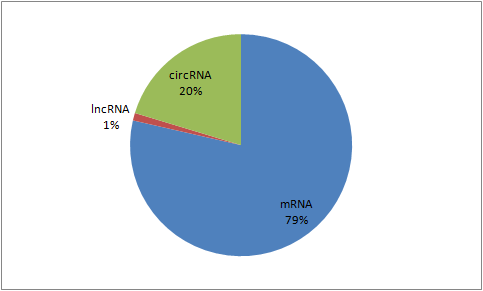
Somatic mutations on predicted miRNA targets.
C. Predicted mRNA-miRNA target sites
We collected somatic mutations within 3' UTRs of RefSeq genes in the hg38 genome from COSMIC. We then assessed if the mutation leads to the creation, disruption, or modification of putative miRNA target sites. We used three methods to predict how the mutations impact miRNA target sites. First, we identified all 3' UTR somatic mutations that create, disrupt, or modify a 6mer or longer sequence complementary to a miRNA seed, using the 6 classes of miRNA seeds described by Ellwanger et al (PMID: 2144577). Second, we used TargetScan to calculate the change in the context+ score for miRNAs binding to the normal and mutated 3' UTR sequence. Context+ scores predict the binding of a miRNA to the entire 3' UTR by summing over contributions made by individual sites within the 3' UTR that have perfect sequence complementarity to the miRNA seed region. A more negative context+ score indicates an increased likelihood for the miRNA targeting the 3' UTR, and, therefore, somatic mutation that increase context+ scores are likely to disrupt miRNA targeting and somatic mutation that decrease context+ scores are likely to increase miRNA targeting. Third, we used PITA to calculate the target score changes for miRNAs binding to the normal and mutated 3' UTR sequence. PITA calculates a target score from free binding energy required for miRNA binding. A higher PITA score indicates higher specificity for miRNA binding. By default, only the impacts of somatic mutations on target sites that are identified by TargetScan are shown. The impact of somatic mutations on all 6mer or longer seed matches can be displayed by clicking the "All 6mer or longer target sites" button on the top of the page. The PITA target scores are labeled as "PITA score change" in the table. We also used the 46-way Multiz alignment of vertebrate genomes to determine if the target site sequence containing the somatic mutation was conserved across species.
D. Predicted lncRNA-miRNA target sites
We downloaded all the lncRNA transcript sequences in FASTA format and their genomic intervals in BED format from LNCipedia. Somatic mutations in lncRNA transcript sequences were then collected from COSMIC. We identified the miRNA target sites that are created, disrupted, or modified in all the 15 base long sequences of lncRNA transcripts with somatic mutations in the middle. For target prediction, first, we applied the 6mer or longer sequence complementary, described by Ellwanger et al (PMID: 2144577). Second, we applied TargetScan prediction. By default, only the impacts of somatic mutations on target sites that are identified by TargetScan are shown. The impact of somatic mutations on all 6mer or longer seed matches can be displayed by clicking the "All 6mer or longer target sites" button on the top of the page.
E. Predicted circRNA-miRNA target sites
circRNA sequences and genomic intervals of circRNA transcripts were downloaded from circBase. Somatic mutations in circRNA transcripts were then collected from COSMIC v73. We identified the miRNA target sites that are created, disrupted, or modified in all the 15 base long sequences of circRNA transcripts with somatic mutations in the middle. For target prediction, first, we applied the 6mer or longer sequence complementary, described by Ellwanger et al (PMID: 2144577). Second, we applied TargetScan prediction. By default, only the impacts of somatic mutations on target sites that are identified by TargetScan are shown. The impact of somatic mutations on all 6mer or longer seed matches can be displayed by clicking the "All 6mer or longer target sites" button on the top of the page.
F. Germline mutations in miRNA target sites
To display germline mutations that could have a similar impact on miRNA targeting as the somatic mutations, we linked the database with data available in PolymiRTS 3.0. Specifically, we search the records available in PolymiRTS for each gene to collect germline mutations that impact target sites of the same miRNAs that are impacted by somatic mutations.
G. GWAS associations with cancer
A table displaying genome-wide association studies collected from the NHGRI GWAS Catalog that have linked the gene with cancer.
H. Candidate gene associations with cancer
Candidate gene association studies collected from the Cancer GAMAdb that have linked the gene with cancer.
I. KEGG pathways
Pathways in the KEGG pathway database in which the gene is a member. Clicking on the Pathway ID provides a list of all genes in the pathway with known somatic mutations that may impact miRNA target sites.
J. Genome Browser Track
A Genome Browser track showing the somatic mutations and putative miRNA binding sites. Two custom tracks are shown: the first displays somatic and germline mutations that alter miRNA target sites and the second displays the putative binding sites that are impacted by the mutations.
Somatic mutations in miRNA sequences
We collected the genomic coordinates of the pre- and mature miRNA sequences from miRBase. The genomic coordinates of miRNAs are compared against the location of somatic mutations from none coding regions. The selected somatic mutations are classified for their positions in the seed, mature and pre-miRNA sequences.
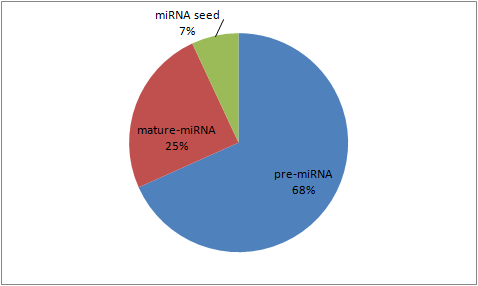
Somatic mutations in miRNA sequences.
For annotating and displaying the functional effects, the seed mutations are linked to miR2GO , our recently published web based tool for comparative functional analysis for microRNAs. miR2GO runs with the default parameter setting to scores the seed mutations in a continuous scale of 0 to 1 for quantifying the semantic changes among the enriched gene ontology terms from target gene sets. A lower score value indicates higher functional impacts and vice versa.
| Column Label | Description |
|---|---|
| miR ID | The miRNA whose targeting is altered by the somatic mutation. Clicking on the miRNA name goes to the description of the miRNA in miRBase. |
| miRNA | Targeting miRNA family name. |
| Cancer Type | The type of cancer in which the somatic mutation was identified. |
| Sample Name | The sample name in which the somatic mutation was identified. |
| miRSite | The sequences containing the somatic mutation. The sequence complementary to the miRNA seed is shown in capital letters. |
| Conservation | The number of vertabrate species in which the miRNA target site sequence is conserved. Clicking on the number provides a list of the species containing the conserved sequence. |
| miRSeed | The sequence of the miRNA seed. |
| SeedClass | The type of seed match that is altered by the somatic mutation of the seed types provided by Ellwanger et al (PMID: 2144577). |
| FuncClass | Whether the somatic mutation creates, disrupts, or modifies the seed match. Mutations that modify seed matches (e.g., 6mer matches become 7mer matches or 8mer matches become 7mer matches) are indicated by the modification of the seed class in parentheses. |
| context+ score change | Difference in TargetScan context+ score between the reference and mutant allele. A more negative value of the context+ score difference indicates an increased likelihood that miRNA targeting is disrupted or newly created by the mutation. | Wildtype CS+ | The context+ score for the miRNA targeting the 3' UTR with the normal allele. "no TS" indicates that the 3' UTR sequence does not contain any target sites for the miRNA that are identified by TargetScan 6.0. |
| Mutant CS+ | The context+ score for the miRNA targeting the 3' UTR with the mutant allele. "no TS" indicates that the 3' UTR sequence does not contain any target sites for the miRNA that are identified by TargetScan 6.0. |
| PITA score change | PITA target score difference for the miRNA targeting the 3' UTR with the mutant and normal allele. Clicking on the number provides PITA scores for normal and mutant allele separately. |
| Location | Genomic location of the mutation. |
| SNP ID | Link to dbSNP. |
| Ancestral Allele | If applicable, the ancestral allele is denoted. |
| Allele | The two possible alleles of the SNP in the mRNA transcript. |
| Validation | Experimental support that the miRNA target the gene. "N" indicates that no experimental support is known. |
| Strand | Directions of transcription. |
| Region | Whether the somatic mutation is located within the precursor miRNA sequence, the mature miRNA sequence, or the seed region of the mature miRNA sequence. |
| miR_Mutation Location | Location of the mutation in miRNA. |
| Functional analysis of the somatic mutation with miR2GO |
Link to miR2GO. Load sequence and mutation from database to miR2GO. Results will be ready approximately in 3 minutes. |
| Mutation | Mutation represents an unique id for somatic mutation in form of Chromosome:g.LocationReferenceAllele>DerivedAllele. |
| Mutation ID | Unique identifier for somatic mutations from COSMIC. |
| Cluster ID | Cluster ID for identifying the associated CLASH, PAR-CLIP, HITS-CLIP experiments. |
| CLASH Sequence ID | Unique ID of interaction from CLASH dataset. |
| Site Type | Type of seed match in a CLASH target site. |
| P-value | P-value of the significance of the association between the disease and mutation. |
| PubMed ID | Link to the PubMed entry describing the study. |
| Experiment Type | CLIP-seq experiment types. |
| RBP | RNA binding protein name. |
| Cell | Name of the cell line. |
| Treatment | Description of the treatment used in the cell line. |
| Dataset | Dataset identifier in the public repositories. |
| SomamiRs | Link to the predicted somamirs in the experimentally identified target sites. |
Cancer Classification
To define cancer types, we adopted the hierarchical classification system from the COSMIC Cancer Browser.
This classification system has four levels: tissue selection, sub-tissue selection, histology selection and sub-histology selection.
For example, the cancer type [haematopoietic_and_lymphoid_tissue][lymph_node][lymphoid_neoplasm][Hodgkin_lymphoma] indicates that the tissue is
"haematopoietic and lymphoid" and the sub-tissue is "lymph node", and the histology and sub-histology are "lymphoid neoplasm" and "Hodgkin lymphoma" respectively. A lis of cancer types and number of somatic mutation records for each cancer type is available for download from here.
Browse
Different browse options allow users to browse records based on target ceRNA types (ALL, mRNA and lncRNA), start letters of gene symbols and cancer types. Following are different browse functions that are available from home page.
A. "Somatic mutations in miRNA sequences" displays somatic mutations in miRNA sequences. The Cancer Type selection allows browsing cancer type specific somatic mutations in miRNA sequences. Clicking on the link "Functional analysis of the somatic mutation with miR2GO" at the last column will load the somatic mutation and miRNA sequence to miR2GO.
B. "Somatic mutations in experimentally identified miRNA target sites: CLASH" displays the somatic mutations in miRNA target sites from CLASH data (Described in 2.A.).
C. "Somatic mutations in experimentally identified miRNA target sites: PAR-CLIP and HITS-CLIP" displays a table of 34 PAR-CLIP and HITS-CLIP experiments. Clicking on the link "Browse Table by Cluster Tag" at the last column will open a second browse page for displaying the records of the corresponding PAR-CLIP or HITS-CLIP data set. Clicking on transcript ids in the second browse page will open the detail record page (Described in 2.B.).
D. "Somatic mutations in predicted miRNA target sites" displays somatic mutations within 3' UTRs of genes, lncRNAs and circRNAs that create or disrupt putative miRNA target sites. Clicking on the transcript id will open the detail record page (Described in 2.C, 2.D and 2.E.).
E. "Biological pathways impacted by somatic mutations in miRNA target sites" displays genes in cancer related KEGG pathways that contain somatic mutations in miRNA target sites. This table provides KEGG pathways containing genes with somatic mutations that impact miRNA target sites. The KEGG pathways are separated into three tables. The first table contains cancer pathways or pathways directly related to cancer pathology, the second section contains signaling pathways, and the third section contains other KEGG pathways. Clicking on the Pathway ID provides the pathway with genes containing 3' UTR somatic mutations in pink boxes as well as a list of these genes with links to the full database record for the gene.
F. "Genes associated with cancer risk that contain miRNA related somatic mutations" displays cancer-associated genes from GWAS and CGAS and also contain somatic mutations in miRNA target sites. Clicking on the gene symbol will open a second browse page for listing all transcript of the gene. Clicking on the transcript symbol will open the detail record page (Described in 2.C, 2.D and 2.E.).
Search
The user can query the database by chromosome location, miRNA ID, Transcript ID, and gene symbol. Clicking on "Example" will load example for selected search option. Clicking on "Submit" will run search query and display results.
Download
Tab-delimited files containing the database contents are available for download here.
miRNA_somatic_v2.0.txt.gz contains somatic mutations in miRNA sequences,
clash_target_somatic_v2.0.txt.gz contains somatic mutations in miRNA target sites identified by CLASH experiments,
clip-seq_somatic_v2.0.txt.gz contains somatic mutations in miRNA target sites identified by PAR-CLIP and HITS-CLIP experiments,
predicted_mRNA_targets_somamir_v2.0.txt.tar.gz contains somatic mutations in predicted miRNA target sites,
lncRNA_somatic_v2.0.txt.gz contains somatic mutations in miRNA target sites on lncRNAs,
circRNA_somatic_v2.0.txt.gz contains somatic mutations in miRNA target sites on circRNAs.
| Please send questions and comments to Dr. Yan Cui at University of Tennesee Health Science Center. |