miR2GO: Comparative functional analysis for microRNAs
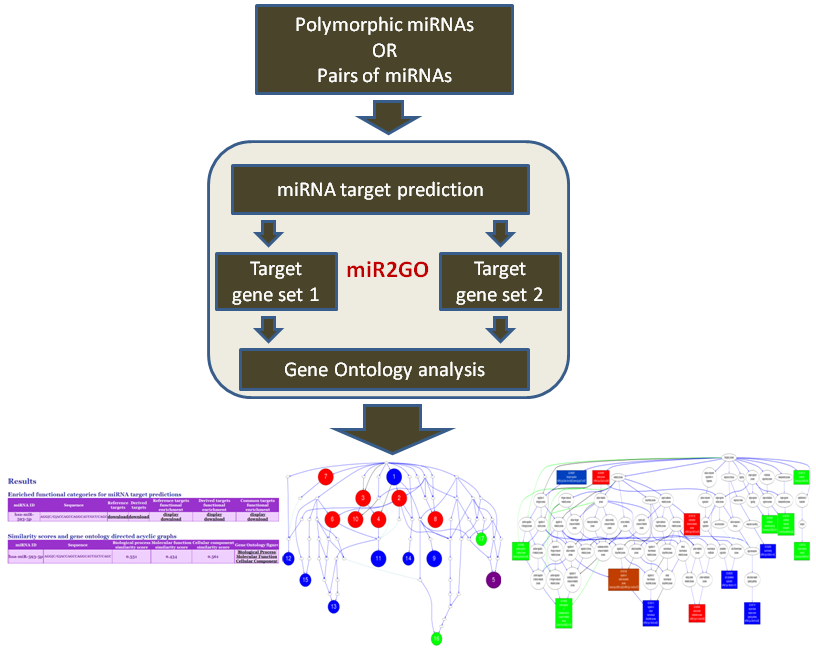
MicroRNAs (miRNAs) are a class of posttranscriptional regulators that involve in almost every biological process. The dysfunctions of miRNAs have been linked with many diseases. A miRNA may interact with Argonaute family proteins to form a RNA-induced silencing complex (RISC) and guide the RISC to bind to mRNA targets. The sequence complementarity between miRNAs and their binding sites in mRNAs is crucial to the target recognition. Genetic and somatic mutations in miRNA sequences, especially those in the seed region, can disrupt the interactions between miRNAs and their targets. These mutations can also make the miRNA bind to a set of new targets. Thus, the mutations in miRNAs can alter miRNA targeting and rewire the miRNA regulatory networks. The main purpose of this web server is to provide a computational tool for the assessment of functional impacts of genetic and somatic mutations in miRNAs. The first function of this web server, miRmut2GO, allows users to analyze the changes of target genes caused by miRNA mutations and view the functional impacts of these changes in a gene ontology graph. This web server can also be used for analyzing the differences and similarities between the functions of different miRNAs. Different miRNAs perform different functions because they bind to different sets of target genes. The second function of the web server, miRpair2GO, allows users to perform comparative gene ontology analysis for the target gene sets of different miRNAs.
*A note about web browser usage: The function of displaying GO figures is not supported by the current version of Internet Explorer. The default threshold for connection timeout in Firefox is 300 seconds. It may take longer time to run the queries containing more multiple mutations. Here is an instruction on how to change the connection timeout setting in Firefox. We suggest to use Google Chrome when accessing miR2GO.
How to cite miR2GO:
Bhattacharya A and Cui Y (2015) miR2GO: Comparative functional analysis for microRNAs. Bioinformatics. btv140
Developed and maintained by: Yan Cui's Lab at University of Tennessee Health Science Center