Microarray Data Analysis I
Yan Cui (ycui2@utmem.edu)
The address of this webpage:
http://compbio.utmem.edu/microarray/lecture1.htm
or
http://compbio1.utmem.edu/microarray/lecture1.htm
Topics:
1.
Hierarchical clustering.
2.
K-Means / K-Medians clustering.
3.
A software for microarray data analysis.
What is Microarray?
Microarray is array of DNA molecules that permit many hybridization experiments to be performed in parallel. It can monitor expression levels of thousands of genes simultaneously.
Microarray has become a powerful tool for biomedical
research. The number of published papers referring to microarray has increased very
fast in recent
years.
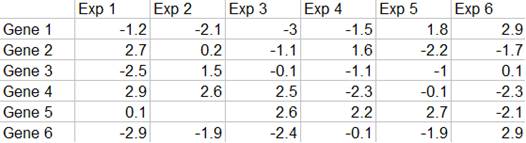
Microarray Data Matrix
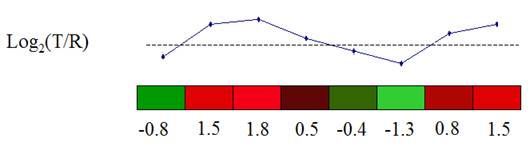
Microarray data are usually presented in an expression matrix. Each column represents all the gene expression levels from a single experiment, and each row represents the expression of a gene across all experiments. Each element is a log ratio. The log ratio is defined as log2 (T/R), where T is the gene expression level in the testing sample, R is the gene expression level in the reference sample.
The expression matrix can be presented as a matrix of colored rectangles. Each rectangle represents an element of the expression matrix.
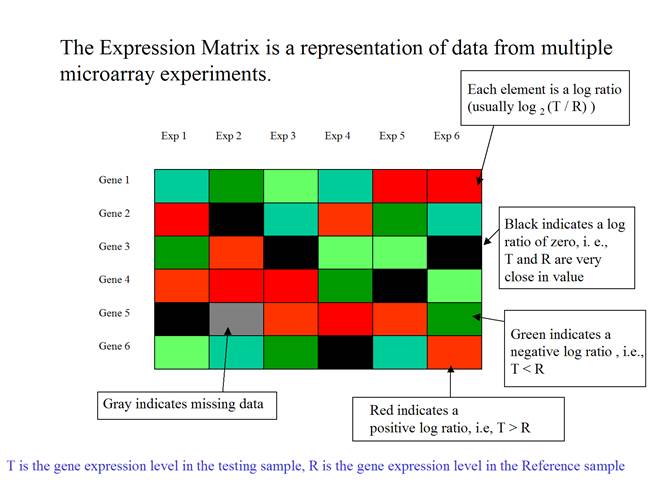
(Adapted from the documentation of MeV, http://www.tigr.org/software/tm4/mev.html)
Hierarchical Clustering
Hierarchical Clustering is the most popular method for microarray data analysis. In hierarchical clustering, genes with similar expression patterns are grouped together and are connected by a series of “branches”, which is called dendrogram (or clustering tree). Experiments with similar expression profiles can also be grouped together using the same method.
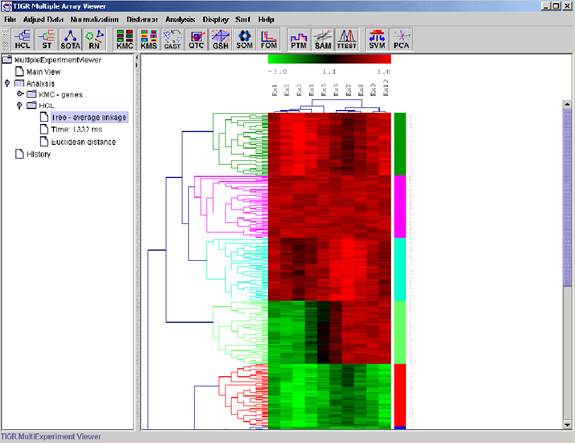
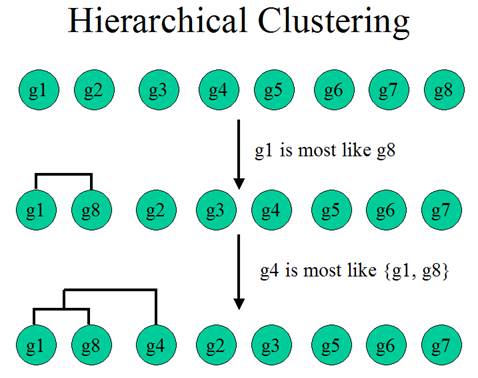
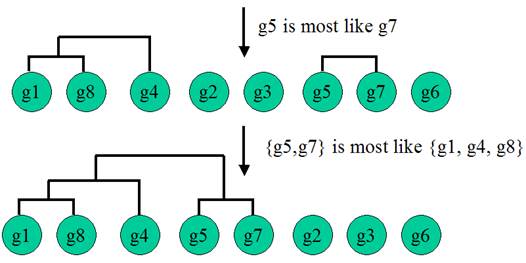
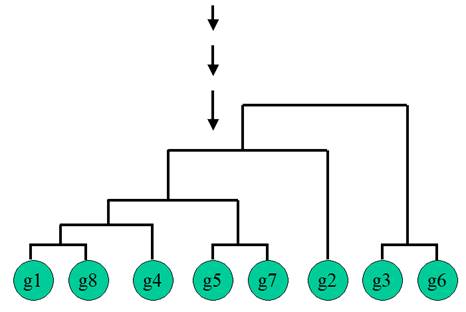
(Adapted from the documentation of MeV, http://www.tigr.org/software/tm4/mev.html)
The
two fundamental problems in hierarchical clustering are:
1.
How
to determine the similarity between two genes?
2.
How
to determine the similarity between two clusters?
To
solve the first problem, we calculate the distance between two expression
vectors. The distance is used as a measure of similarity between genes.
A Gene Expression Vector consists of the expression of a gene over a set of experimental conditions.
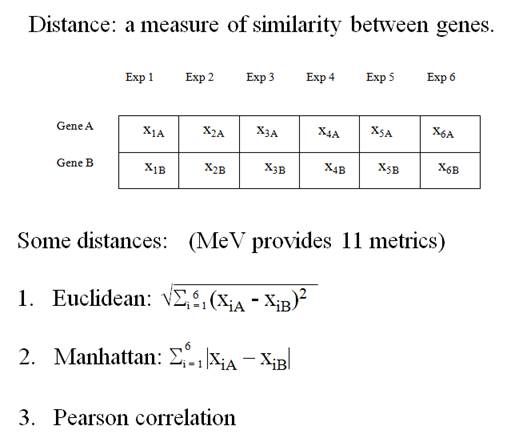
(From the documentation of MeV, http://www.tigr.org/software/tm4/mev.html)
The
second problem is: How to determine the similarity between clusters? The method
for determining cluster-to-cluster distance is called linkage method.
Three
linkage methods:
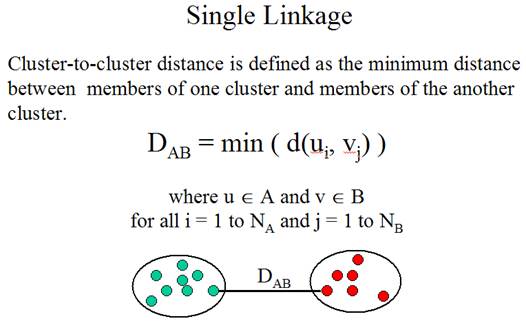
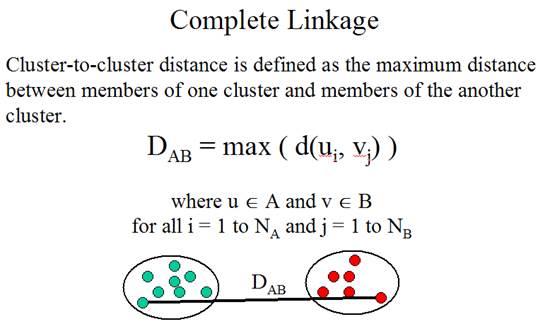
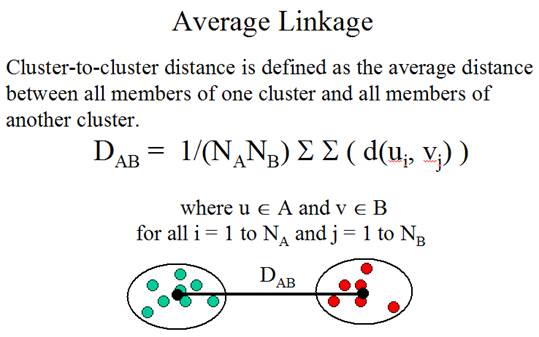
(Adapted from the documentation of MeV, http://www.tigr.org/software/tm4/mev.html)
There
is no guideline for selecting the best linkage method. In practice, people
almost always use average linkage.
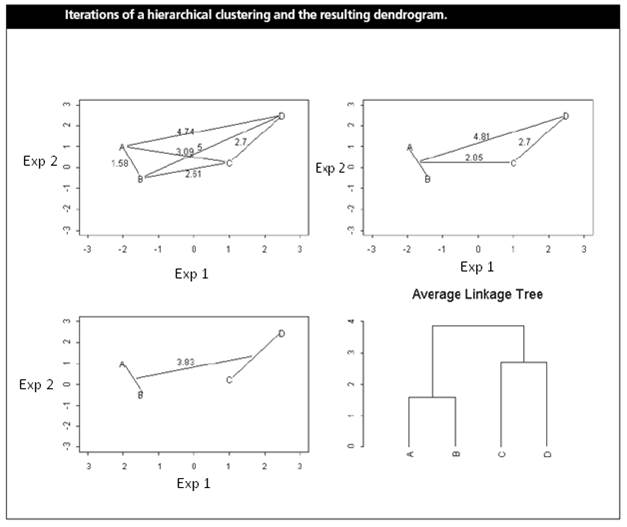
(Shannon W. et
al. (2003) Analyzing microarray
data using cluster analysis. Pharmacogenomics 4:41-51.)
Gene A and B are merged first at the level of 1.58.
The position of the “splitting point” shows the distance (or similarity)
between two genes (or clusters). A low “splitting point” indicates high
similarity.
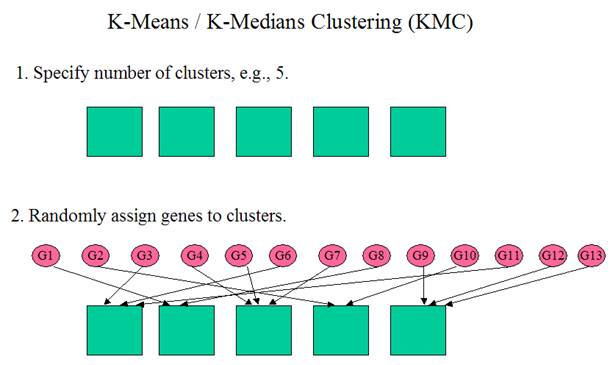
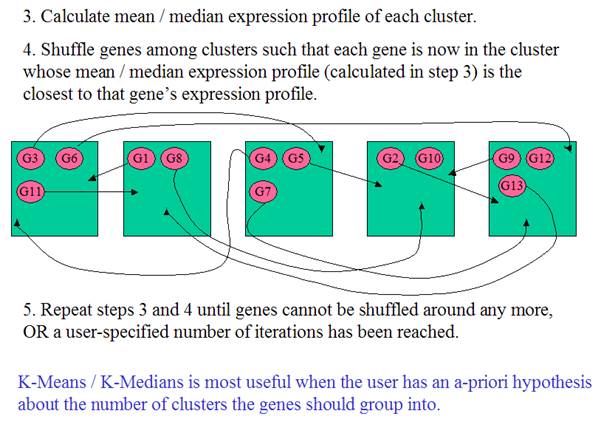
K-Means / K-Medians Clustering
What is Mean? What is
Median?
Mean
is the average.
Median
is the “middle number”, i.e. the middle of the distribution. When there is an
odd number of numbers, the median is simply the middle number. For example, the
median of 2, 4 and 7 is 4.
When
there is an even number of numbers, the median is the mean of the two middle
numbers. Thus, the median of the numbers 2, 4, 7, 12 is (4+7)/2 = 5.5.
Median is more robust against outliers
For example, the mean of 5, 6, 7, 8 and 9 is 7. The
median of 5, 6, 7, 8 and 9 is also 7.
But, the mean of 5, 6, 7, 8 and 99 is 25, while the
median of 5, 6, 7, 8 and 99 is still 7.
(From
the documentation of MeV, http://www.tigr.org/software/tm4/mev.html)
TIGR MultiExperiment Viewer
TIGR
MultiExperiment Viewer (MeV) is a software for microarray data analysis.
MeV was developed by a group of people at TIGR (The Institute for Genomic
Research) and is freely available from TIGR
web site. Mev uses The Artistic
License, which means you can freely download the software or get a copy
from another user (for details see http://www.tigr.org/software/tm4/generalFAQ.html).
Running MeV requires Java Runtime Environment (JRE).
A detailed instruction on installation of JRE and MeV can be found at http://compbio.utmem.edu/MSCI814/faq.php.
Starting TIGR MultiExperiment Viewer
The program is in
C:\TMEV. Double
click the batch file (TMEV.bat) to start the program. Use the File menu
to open a new Multiple Array Viewer.
Loading Microarray Data
Select Load data from the File menu to
launch the file-loading dialog. At the top of this dialog, use the drop-down
menu to select the type of expression files to load. Use the file browser to
locate the files to be loaded.
HCL: Hierarchical clustering
Type of the Input file: Stanford Files (*.txt)
Name of the Input file: Stanford_Large.txt
Select Options:
1.
Average
Linkage;
2.
Cluster
both genes and experiments.
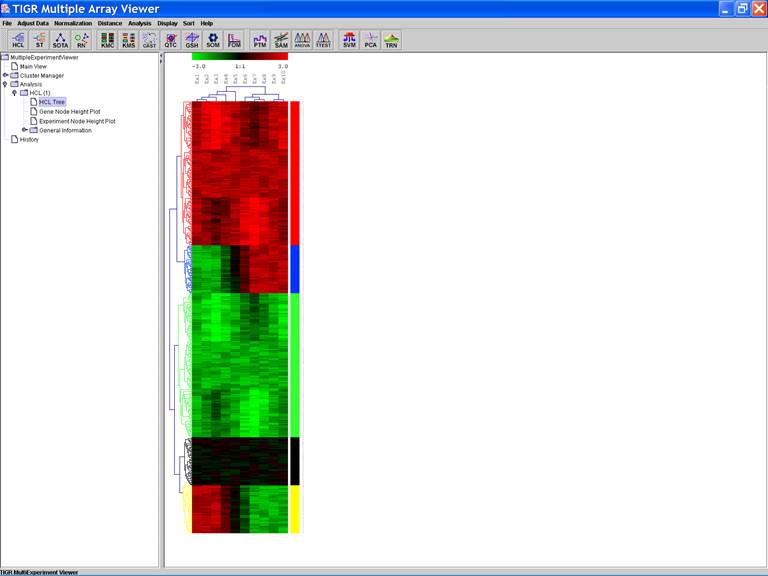
Working with clusters
Clusters of interest can be stored:
1.
Click
the dendrogram to select the cluster;
2.
Open
a menu by right clicking in the viewer and selecting the store cluster
option;
3.
Input
the name of the cluster and select a color to label the cluster.
A color bar is displayed along the right side of
cluster.
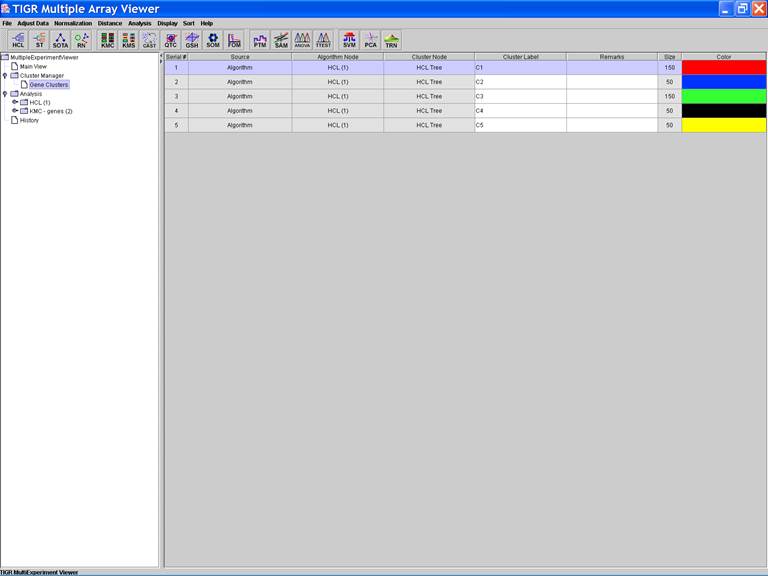
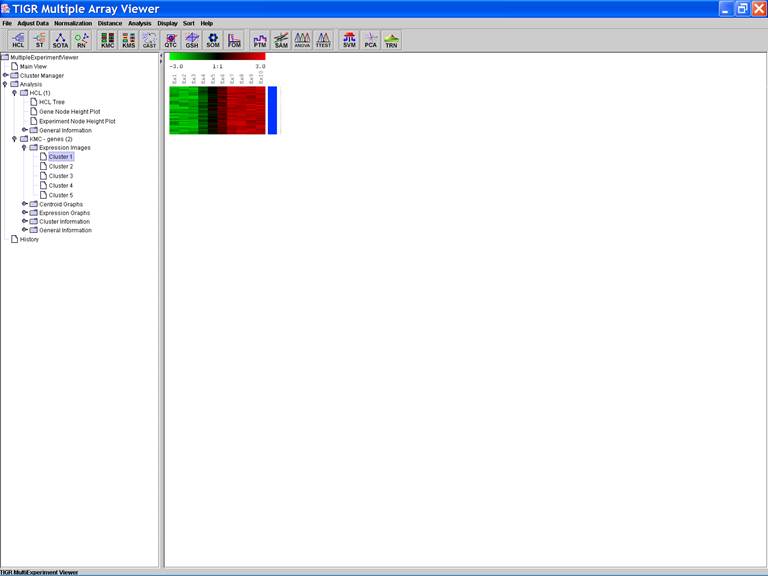
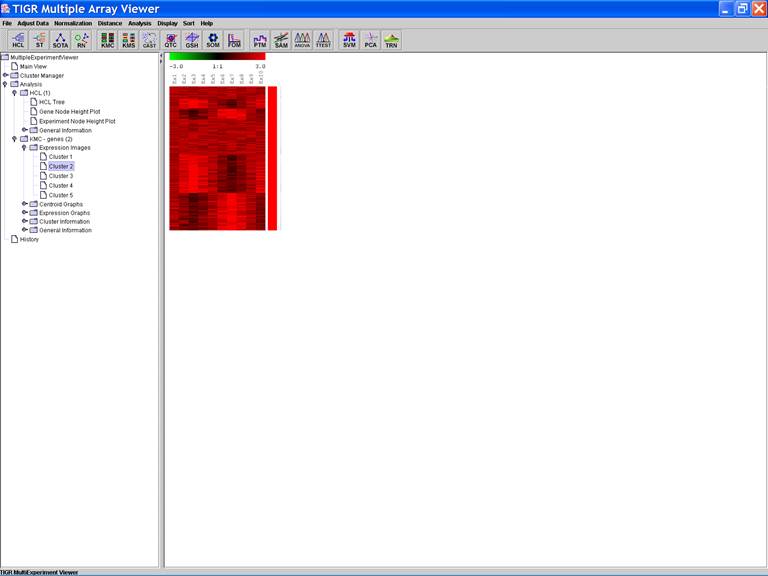
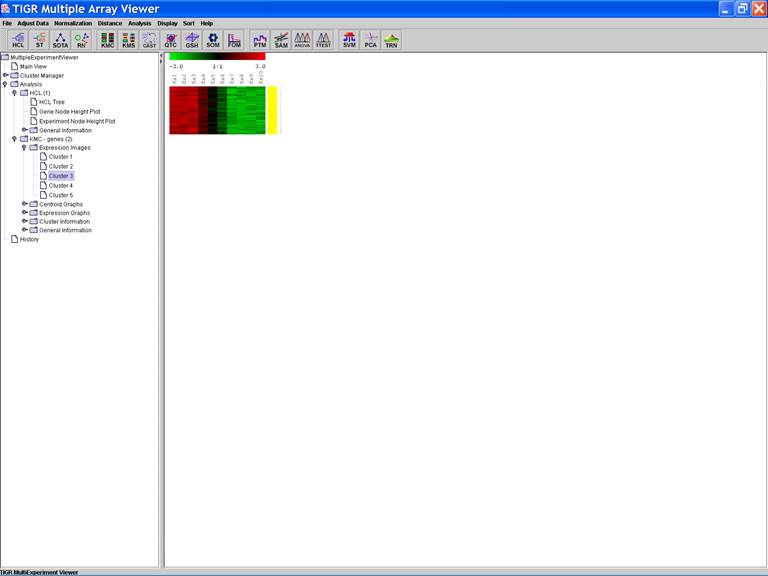
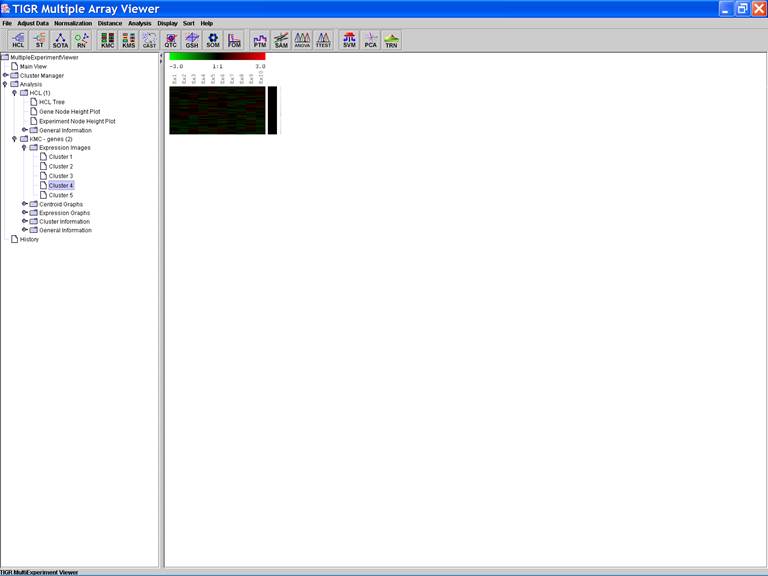
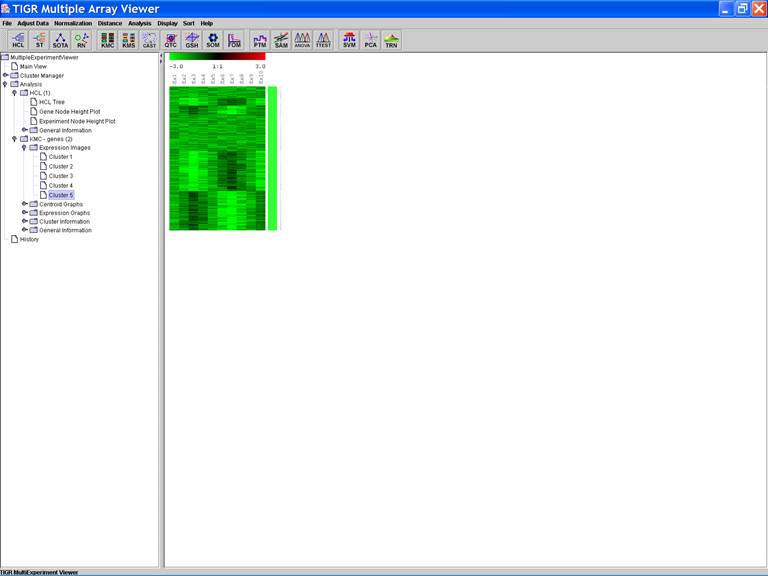
KMC: K-Means / K-Medians Clustering
Options:
1.
Cluster
genes;
2.
Use
mean;
3.
Number
of clusters = 5;
4.
Number
of iterations = 50;
Further Reading
1.
The
review articles on microarray analysis in the
three special
issues of
Nature
Genetics
a.
The
Chipping Forecast I (http://www.nature.com/ng/journal/v21/n1s/index.html)
b.
The
Chipping Forecast II (http://www.nature.com/ng/journal/v32/n4s/index.html)
c.
The
Chipping Forecast III (http://www.nature.com/ng/journal/v37/n6s/index.html)
2.
Shannon
W. et al. (2003) Analyzing microarray data using cluster analysis.
Pharmacogenomics 4:41-51. (http://ilya.wustl.edu/~shannon/pharmacogenomics.pdf)
Homework
Due Date: Wednesday, March 15. Submit to Dr. Yan Cui via
email (ycui2@utmem.edu). The solution will
be posted at http://compbio.utmem.edu/MSCI814/Solution1.htm
on March 16.
Background: DNA microarray has shown great promise in studying
complex diseases such as cancer. The genome-wide gene expression profiles of tumor
tissues are considered as the “molecular portraits” of various cancers. For
example, Clustering
of breast and ovarian carcinoma cases is shown in the figure below, 68 breast
and 57 ovarian cases were co-clustered to discern both similarities and disparities
between the two sample sets. The common reference control consisting of equal
amounts of mRNA from 11 human cancer cell lines. (Schaner, M et al., Gene Expression
Patterns in Ovarian Carcinomas, Mol Biol Cell. 2003
Nov;14(11):4376-86).
Data: Download the dataset from http://compbio.utmem.edu/MSCI814/Homework1.txt
(Right click on the link and select “Save Target As…”). The
dataset contains gene expression profiles of 16 tumor samples. Each
of the 16 samples is
associated with one of the two cancers.
1. Analyze
the data with hierarchical clustering (HCL)
You should use average linkage method, Euclidean
distance metric and only cluster experiments.
Please infer from the dendrogram (clustering tree) the
two groups of the samples (each associated with a type of cancer).
For example,
Cancer 1: Sample 1, 3,5,7,9,11,13,15
Cancer 2: Sample 2,4,6,8,10,12,14,16
2. Analyze the data with
K-means clustering (KMC)
Use K-means clustering method to group the 16
samples into two clusters.
For example,
Cluster 1: Sample 1, 3,5,7,9,11,13,15
Cluster 2: Sample 2,4,6,8,10,12,14,16
What should be included in the email:
1. HCL
Cancer 1: Sample…
Cancer 2: Sample…
2. KMC
Cluster 1: Sample…
Cluster 2: Sample…